Afrocentric Social Science Revolutions that Changed the Africalogical World View
Considerable progress has been made in the africalogical research of ancient history (DuBois, 1965, 1970; Diop, 1974,1991; Winters 1981\1982, 1991, 1994), classical studies (Parker, 1917, 1918) and the role of Blacks in ancient Indo-China and China (Winters, 1985) over the past 90 years. Marcus Garvey (1966) and DuBois (1965,1970) had a tremendous influence in the study of ancient Black history.
.

Marcus Garvey with the founding of the UNIA attracted many africalogical researchers to his organization. Some of these researchers wrote articles for the Negro World newspaper. These scholars formed the foundation for the africalogical scientific community including C.G Woodson, J. A. Rogers, William Ferris, George W. Parker and Arthur A. Schomberg.
In addition to members of the UNIA playing a prominent role in the precision of prediction of the africalogical paradigm we find that afrocentric researchers belonging to the Hamitic League of the World (HLW), also contributed greatly to the enhancement of the "Ancient Model" and the ASAH paradigms at the turn of the 20th century. John Albert Williams founded the HLW. Two its most prominent members include G. W. Parker (1917,1918) and A.A. Schomberg (1925).
G.W. Parker greatly expanded the ASAH paradigm for classical study by providing a focused study of the role of blacks in Greece. Parker (1917) identified these ancient Afro-Greeks as Pelasgians. He also used linguistics to illustrate that the names of many Greek heroes betrayed there African, not Indo-European origin. In addition, Parker gave us the most detailed discussion of Blacks in India up to his time (Parker, 1918).
The second major confirmation for the "Ancient Model" of history was made by DuBois (1965,1970). In the Negro published in 1915, DuBois explained the African presence in Egypt and ancient Kush and a comprehensive analysis of the West African empires.
W.E.B. DuBois (1924) also firmly placed the presence of Blacks in America as a legitimate research area for africalogical researchers. In The Gift of Black Folks, discussed the Black presence in ancient America, including European references to Pre-Columbian Blacks, and the influence of Africans on the Amerindian religions (DuBois,1924). The confirmation of this paradigm was made by ( Clegg, 1975; Lawrence, 1962; Thompson, 1975; Winters, 1981\1982)
In The World and Africa, DuBois (1965) provides a full explanation of the role of Blacks in the early world. He explains the history of Blacks in China and India (pp.176-200); Blacks in Europe(the Pre-Indo-European Greeks and during the Dark Age of Greece), and Asia Minor (pp. 115-127), and the Egyptian foundation of Grecian thought (pp. 125-126).
The major revolution in the ASAH was the research of Diop (1974, 1991). Diop, a Senegalese expert on Egyptian and African history made important contributions to the ASAH paradigms, including:
1) clarification of the African role in Egypt;
2) proved positively that the West African people formerly lived in close proximity to the Egyptians who were in many cases their ancestors;
3) made Mdu Neter the Egyptian language the classical language for ancient africalogical research; and
4) developed the genetic model as the major paradigm in the ASAH (Diop, 1991).
In general, Diop (1974, 1991) caused an africalogical social scientific revolution because he was able to prove that Egypt was the archetypical civilization for many West Africans. This was an important discovery because almost all of the slaves that were sold in the United States had originally came from West Africa. Verification of the Egyptian origin of West Africans provided African Americans with relationship to the ancient Egyptians.
Moreover, Diop's use of linguistics, and anthropological evidence to confirm the African origin of Egypt eliminated the need for africalogical researchers to use the classical writers to prove the African origin of Egypt (Diop, 1977, 1978, 1981, 1986, 1987, 1988). This finding by Diop has led africalogical researchers to seek a better understanding of African philosophy through an interpretation of Egyptian philosophy. Moreover, africalogical researchers have also began the reconstruction of the Paleo-African language used by Blacks in prehistoric times (Anselin, 1982, 1982b, 1989; Winters, 1994) so that we will know more about the culture and civilization of the Proto-Africans.
The last major confirmation of the ASAH paradigms was made by Clyde Ahmad Winters (1977, 1979, 1981, 1983a, 1983c, 1983d, 1984, 1985) when he expanded our understanding of the role of Blacks\Africans in Indo-China, India and China; and the ancient literacy of Blacks (1979, 1983d, 1985c, 1986b). The major work of Dr. Winters is Afrocentrism: Myth or Science .
To conduct Afrocentric historical and anthropological research Dr. Winters uses the genetic model of historical research. Dr. Winters outlines these methods in his article “ Ancient Afrocentric History and the Genetic Model”, in Egypt and Greece, edited by Molefi Kete Asante and Ama Mazama
.

.
C. Anta Diop is the founder of modern Afrocentricism . Diop (1974,1991) laid the foundations for the Afrocentric idea in education. Diop (1974, 1991) has argued that the genetic model can be used to explain the analogy between ancient African civilizations. There are three components in the genetic model: 1) common physical type, 2) common cultural patterns and 3) genetically related languages (Winters 1989a). Diop over the years has brought to bear all three of these components in his illumination of Kemetic civilization (Diop 1974,1977,1978,1991). Dr. Winters took these components of the genetic model to explore the history of Black and African civilizations around the world.
Gates, like most Afro-Americans in the Academe are afraid to present an Afrocentric view of African and Black history for fear of losing their jobs or being ostracized by white colleagues.
Many Eurocentrist believe that African-Americans should only write about slavery and leave the writing of ancient history to more "qualified" scholars. Moitt (l989) observed that:"The limitation has come about of the bias in historiography. The central problem is that historians have made plantation slavery and its effects in the Americas their sole preoccupation. And they have persuaded their students to do likewise. The damage this has done is incalculable. Blacks viewed their history and, by extension that of Africa in terms of slavery".(p.358) This is why Gates concentrates in the Great Empires of Africa, of civilizations influenced or recognized by Europeans, instead of using the archaeological evidence to influence his review of African history.
Moitt (l989) believes that this desire to deny blacks a role in ancient history is the root cause of white opposition to Diop. He wrote that: "All of this raises the question of historical methodology and goes to the heart of the matter of Diop's isolation....To what must we attribute this negation of Diop? The negation goes beyond the artificial division of the African continent at the Sahara desert and hinges on Diop's ideas the most contentious of which is that the ancient Egyptians were Blacks. In this respect, the negation is not of Diop alone, but formation, Bantu migration, Islam and the slave trade are seen as major problems in African history, the debate over Egypt is stifled" (Moitt 1989,p.358).
Dr. Winters using linguistic, anthropological and historical evidence, he proved that the earliest cultures of China and Indo-China were founded by Blacks from West Africa and modern Ethiopia (Winters, 1979, 1983d, 1985c, 1986b). The Black Civilizations of Asia are also discussed in this book The Ancient Black Civilizations of Asia.

Winters also made it clear that the earliest Japanese were Blacks and that Japanese is related to Mande/African Languages (Winters, 1979, 1981, 1983a, 1983c, 1984). In addition he was able to prove that the founders of Xia and Shang were of African and Dravidian origin (1983c,1985c).


Using the findings of Wiener in regards to the writing of the Olmecs Winters discovered that the Blacks from West Africa left numerous inscriptions written in the Manding language (Winters, 1977, 1979, 1983a, 1985b) . Dr. Winters explains the Olmec-Mande connections in [b]Atlantis in Mexico and African Empires in Ancient America.
Winters later discovered that due to the cognition between the Mande writing and ancient scripts used by the Minoans and Indus Valley he could read the Indus Valley Writing and the Linear A inscriptions (1985b). Dr. Winters has written a detailed grammar and dictionary of the Indus Valley Writing and the Mande origin of the Minoan Linear A .


This africalogical research by Winters (1981/1982, 1983b, 1983d, 1989a, 1991, 1994) made it clear that the first civilizations in Indo-China and China were founded by Blacks. He has also proved the lie to Hume's (1875) claim that Blacks have "No literacy" and "No letters".
Today genetics research is used to support population migrations. Dr. Winters has began to publish numerous articles on on the genetic history of Black and African people.

REFERENCES
Anselin, A. (1982). Le mythe d' Europe. Paris: Editions Anthropos.
_______.(1982b). "Zeus, Ethiopien Minos Tamoul", Carbet Revue Martinique de Sciences Humaines,no. 2:31-50.
_______.(1989). "Le Lecon Dravidienne",Carbet Revue Martinique de Sciences Humaines, no.9:7-58.
Asante,M.A. (July-August, 1996). "Ancient Truths", Emerge , 66-70.
Asante,M.K. (1990) Kemet,Afrocentricity,and Knowledge. Trenton ,NJ:Africa World Press.
_________ (1991). "The Afrocentric idea in Education",Journal of Negro Education,60(2):170-180.
__________.(December 1991/January 1992). "Afrocentric Curriculum".Educational Leadership, pp.28-31.
Bernal, M. (1996, Spring). The Afrocentric interpretation of history: Bernal replies to Lefkowitz. Journal of Blacks in Higher Education, 86-95.
Bernal,M. (1987). Black Athena. New York: Free Association Press. Volume 1.
________. (1991). Black Athena. New York: Free Association Press. Volume 2.
Blyden, E.W. ( January, 1869). The Negro in ancient history. Methodist Quarterly Review, 71-93.
Blyden, E.W. (1887). Christianity, Islam and the Negro Race. Edinburgh: Edinburgh University Press.
_____________. (1890). The African Problem and the method for its solution. Washington, D.C.: Gibson Brothers.
_______________.(1905). West Africa before Europe. London: C.M. Phillips.
Clegg, L.H. (1975). Who were the first Americans? The Black Scholar, 7(1), 32-41.
Coleman, B.E. (1971). A history of Swahili, The Black Scholar, 2 (6), 13-25.
Cornish, S. & Russwurm, J.B. (1827). European colonies in America, Freedom Journal, 1.
Carruthers, J. (1977). Writing for Eternity, black book bulletin, 5 (2), 32-35.
Carruthers, J. (1980). Reflections on the history of afrocentric worldview, black book bulletin, 7(1), 4-13, 25.
Delany, M.R. (1978). The origin of races and color. Baltimore, M.D.: Black Classic Press.
Diop,C.A. (1974). The African Origin of Civilization. (ed. & Trans) by Mercer Cook, Westport:Lawrence Hill & Company.
_________.(1977). Parente genetique de l'Egyptien Pharaonique et des Languaes Negro-Africaines. Dakar: IFAN ,Les Nouvelles
Editions Africaines.
__________.(1978) The Cultural Unity of Black Africa. Chicago: Third World Press.
__________. (1981). A Methodology for the study of migration. UNESCO (Ed.), African Ethnonyms and Toponyms, (pp.87-110). Paris: UNESCO.
___________.(1986). "Formation of the Berber Branch". In Libya Antiqua. (ed.) by Unesco,(Paris: UNESCO) pp.69-73.
____________.(1987). Precolonial Black Africa. (trans. ) by Harold Salemson, Westport: Lawrence Hill & Company.
____________.(1988). Nouvelles recherches sur l'Egyptien ancient et les langues Negro-Africaines Modernes. Paris: Presence
Africaine.
_____________(1991). Civilization or Barbarism: An Authentic Anthropology. (trans.) by Yaa-Lengi Meema Ngemi and (ed.) by H.J. Salemson and Marjoliiw de Jager, Westport:Lawrence
Hill and Company.
Douglas, F. (1966). The claims of the Negro ethnologically considered. In H. Brotz (Ed.), Negro social and political thought (pp. 226-244). New York: Basic Books, Inc., Pub.
DuBois, W.E.B. (1924). The Gift of Black Folks. Boston.
DuBois, W.E.B. (1970). The Negro. New York: Oxford University Press.
DuBois, W.E.B. (1965). The world and Africa. New York : International Publishers Co., Inc.
Ferris, W.H. (1913). The African abroad. 2 vols. New Haven,CT :Tuttle, Morehouse and Taylor.
Garvey, M. (1966). Who and What is a Negro. In H. Brotz (Ed.), Negro social and political thought (pp. 560-562).New York: Basic Books, Inc. Publishers.
Graves, Robert. (1980). The Greek Myths. Middlesex:Peguin Books Ltd. 2 volumes.
Hansberry, L.H. (1981). Africa and Africans: As seen by classical writers (Vol. 2). Washington, D.C.: Howard University Press.
Hopkins, P.E. (1905). A Primer of Facts pertaining to the early greatness of the african race and the possibility of restoration by its descendants-with epilogue. Cambridge: P.E. Hopkins & Com.
Hume, D. (1875). Essays: Moral political and literary. T.H. Green and T.H. Grose. 2 Vols. London.
Jackson, J. (1974). Introduction to African civilization. Secaucus, N.J.: Citadel Press.
James, G.M. (1954). Stolen legacy. New York: Philosophical Library.
Kuhn, T.S. (1996). The structure of scientific revolution. Chicago: University of Chicago Press.
Lacouperie, Terrien de. (1891). The black heads of Babylonia and ancient China, The Babylonian and Oriental Record, 5 (11), 233-246.
Lawrence, H.G. (1962). African explorers of the New World, The Crisis, 321-332.
Merton, R.K. (1957). Social theory aand social structure. Glencoe, Ill. : The Free Press.
Moitt,B. (1989). "Chiekh Anta Diop and the African Diaspora: Historical Continuity and Socio-Cultural Symbolism". Presence Africaine, no. 149-150:347-360.
Parker,G.W. (1917) . "The African Origin of Grecian Civilization ".Journal of Negro History, 2(3):334-344.
___________. (1981). The Children of the Sun. Baltimore,Md.: Black Classic Press.
Perry, R.L. (1893). The Cushite. Brooklyn: The Literary Union.
Rawlinson, George. (1928).The History of Herodutus. New York : Tudor.
Schomburg, A.A. (March, 1925).The Negro digs up his past. Survey Graphic, 670-672.
Schomburg, A.A. (1979). Racial integrity. Baltimore, M.D.: Black Classic Press.
Thompson, Jr. A.A. (1975). Pre-Columbian [African] presence in the Western Hemisphere, Negro History Bulletin, 38 (7), 452-456.
Williams, G.W. (1869). History of the Negro Race in America. New York: G.P. Putnam.
Wimby, D. (1980). The Greco-Roman Tradition concerning Ethiopia and Egypt, black books bulletin, 7(1), 14-19, 25.
Winters, C.A. (1977). The influence of the Mande scripts on ancient American writing systems", Bulletin l'de IFAN, T39, serie B, no. 2 (1977), pp.941-967.
Winters, C.A. (1979). Manding Scripts in the New World", Journal of African Civilizations, l(1), 80-97.
Winters,C.A. (December 1981/ January 1982). Mexico's Black Heritage. The Black Collegian, 76-84.
Winters, C.A. (1983a). "The Ancient Manding Script". In Blacks in Science:Ancient and Modern. (ed.) by Ivan van Sertima, (New Brunswick: Transaction Books) pp.208-215.
__________. (1983b). "Les Fondateurs de la Grece venaient d'Afrique en passant par la Crete". Afrique Histoire (Dakar), no.8:13-18.
_________. (1983c) "Famous Black Greeks Important in the development of Greek Culture". Return to the Source,2(1):8.
________.(1983d). "Blacks in Ancient China, Part 1, The Founders of Xia and Shang", Journal of Black Studies 1 (2), 8-13.
________. (1984a). "Blacks in Europe before the Europeans". Return to the Source, 3(1):26-33.
Winters, C.A. (1984b). Blacks in Ancient America, Colorlines, 3(2), 27-28.
Winters, C.A. (1984c). Africans found first American Civilization , African Monitor, l , pp.16-18.
_________.(1985a). "The Indus Valley Writing and related Scripts of the 3rd Millennium BC". India Past and Present, 2(1):13-19.
__________. (1985b). "The Proto-Culture of the Dravidians, Manding and Sumerians". Tamil Civilization,3(1):1-9.
__________. (1985c). "The Far Eastern Origin of the Tamils", Journal of Tamil Studies , no.27, pp.65-92.
__________.(1986). The Migration Routes of the Proto-Mande. The Mankind Quarterly,27 (1), 77-96.
_________.(1986b). Dravidian Settlements in Ancient Polynesia.India Past and Present, 3 (2), 225-241.
__________. (1988). "Common African and Dravidian Place Name Elements". South Asian Anthropologist, 9(1):33-36.
__________. (1989a). "Tamil, Sumerian, Manding and the Genetic Model". International Journal of Dravidian Linguistics,18(1):98-127.
__________. (1989b). "Review of Dr. Asko Parpola's 'The Coming of the Aryans'", International Journal of Dravidian Linguistics, 18(2):98-127.
__________. (1990). "The Dravido-Harappan Colonization of Central Asia". Central Asiatic Journal, 34(1/2):120-144.
___________. (1991). "The Proto-Sahara". The Dravidian Encyclopaedia, (Trivandrum: International School of Dravidian Linguistics) pp.553-556. Volume l.
----------.(1994). Afrocentrism: A valid frame of reference, Journal of Black Studies, 25 (2), 170-190.
_________.(1994b). The Dravidian and African laguages, International Journal of Dravidian Linguistics, 23 (1), 34-52.
Woodson, C.G. & Wesley, C.H. (1972). The Negro in Our History. Washington, D.C. Associated Publisher.

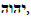 as well as Enoch’s Word of God,
as well as Enoch’s Word of God, 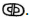 Both “Words” are rooted in the Luwian hieroglyphics for “God” and “Gate” that are carved into G
Both “Words” are rooted in the Luwian hieroglyphics for “God” and “Gate” that are carved into G