The Khoisan are the ancestors of the Black Berbers whoes descendants probably live in Morocco and the Atlas Mountains.
The Black Berbers of the Atlas Mountains and other parts of Northwest Africa are of Sub-Saharan origin and took African mtDNA into Western Europe over 40kya. The Gibraltar Straits appears to be the most reliable route for the spread of many mtDNA haplogroups from Africa, into Europe over the past 30ky (Winters,2012), including L3(M,N) .
The Khoisan carry L1c,L1i, L2b, L3d ( Rito, et al ,2013) . The motif L3b, is widespread in western Africans. It is mainly found among populations that speak languages of the Niger-Congo family like the Mandekan.
Like most African haplogroups the control region of hg L1i include 16189,16223 and 16311, just like L3a and L3b. The mutation that connects the Khoisan to the spread of L3(M,N) is AF24. The AF24 mutation is found in LOd and among the Khoisan and Senegalese .The existence of AF24 in Senegal and Southern Africa suggest that L1c, L2b, L3d and L3e is not the result of intermarriage with Bantu immigrants , as suggested by Rito et al(2013) .
LOd is the oldest mtDNA haplogroup . This haplogroup is primarily carried by the Khoisan people (Winters,2014) . It is also found among Niger-Congo speakers in West Africa where we also find LOa in West Africa in addition to L3b.
The Cro Magnon DNA found in the ancient skeletons dates back to the Aurignacian period (Winters,2011). The Cro magnon skeletons belong to the N haplogroup.
The Cro Magnon skeletons carried N1a,N1b,N1c and N* (Winters, 2010,2011). It is characterized by motifs 00073G,10873C, 10238T and A4CC between nucleotide positions 10397 and 10400. Most of the skeletons carried hg N*.
It is obvious that L3 (M,N) had expanded into Europe prior to the Neolithic.
.
.
Frigi et al (2010), in Ancient Local Evolution of African mtDNA Haplogroups in Tunisian Berber Populations noted that: “Our results also point to a less ancient western African gene flow to Tunisia involving haplogroups L2a and L3b. Thus the sub-Saharan contribution to northern Africa starting from the east would have taken place before the Neolithic. The western African contribution to North Africa should have occurred before the Sahara’s formation (15,000 BP)”.
This would explain why Pericot and Dominguez (2005) found evidence of hg L3 at ancient Iberian sites. Luis Pericot was sure that the populations associated with the Gravettian (32kya) and Soultrean (23kya) cultures were phylogenetically Sub-Saharan African (Dominguez,2005). Dominguez (2005) found that the lineages recovered from ancient skeletons associated with these cultures belonged to the African lineages L1b,L2 and L3. Almost 50% of the lineages from the Abauntz Chalcolithic deposits and Tres Montes, in Navarre are the Sub-Saharan lineages L1b,L2 and L3.
Berber and Khoisan Y-Chromosome
The archaeological data support a migration of probable Khoisan migration from Southern Africa to North Africa. You have to remember that many populations have settled Morocco, so their might not always be a one-to-one correspondence between contemporary Khoisan haplogroups and haplogroups found among contemporary Berbers in Morocco and the Atlas mountains. But given the geography, we would expect to see elements of Khoisan relic population genes among Atlas Mountain Berbers.
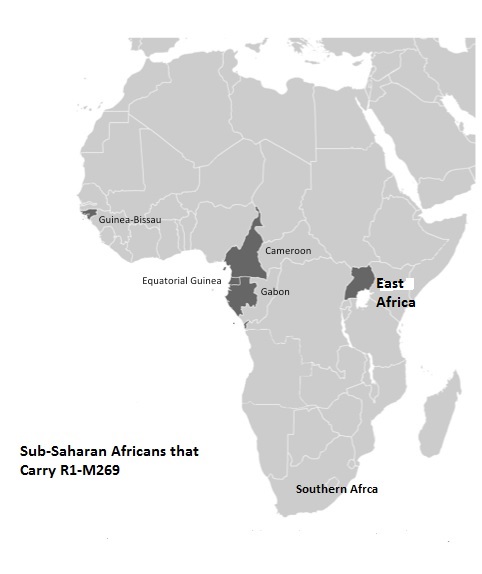
quote:The Berbers and Khoisan also carry similar R1 haplogroups. The Khoisan in South Arica carry R1-M343 (Schlebusch,,2010). Berbers at Zenata and Oran also carry R1-M343 (Bekada, et al, 2010).
Originally posted by Troll Patrol # Ish Gebor:
Figure S1. Map Showing Location of the Population Samples Considered in This Study
Populations are represented by circles and numbered as in Table S5. Sectors within circles are proportional to the frequency of haplogroup A1a (green), A1b (red) and A2-T (black). Green asterisks indicate countries were haplogroup A1a has previously been observed.
See:
Table S1. Haplogroup Affiliation of the Seven Chromosomes that Were Re-sequenced
Table S5: Populations considered for the mutations defining major clades A1b, A1a and A2-T.
http://www.cell.com/cms/attachment/1088206/8032906/mmc1.pdf
The hominids and tool kits common to South Africa also appear in Morocco. It is no secret that the earliest Y-chromosome haplogroups have been found in Morocco and among the Khoisan. These haplogroups belong to hg A (M91); among the Moroccans we find A1b and A1a.The Khoisan mtDNA was named originally L1a,L1d and L1k, these clades are called LoD and LoK today.
Morocco has yielded impotant new data on African prehistory. Here we find a complex and rich set of early hominids from Jebel Irhoud, Dar es-Soltan and Contrebandiers Cave.
A pan-African Middle Stone Age (MSA) culture existed that united South Africa and Morocco.The Moroccan tools are Levallois technology and Mousterian industries were used in South Africa,North Africa and western Eurasia. Dibble et al (2013) has shown that Pan-African industries included cognate scrapers, Levallois tools, Nassarius beads and engraved ostrich shells. The Moroccans and South Africans shared Levallois tools and the use of ochre, bone tools and ostrich shells. Bouaouggar et al (2007) has shown how the shell beads from Grotte des Pigeons (Taforalt,Morocco) and South Africa's Blombos Caves. The archaeological evidence is clear the Khoisan in Morocco and South Africa shared behavioral , cognitive and technological styles. The major behavioral indicators shared by the Moroccan and South African Khoisan was mining,beads, blades, ochre and bone tools between 200-40kya. It is important to note that Moroccan tools are Levallois technologies and Mousterian industries used in North Africa and Western Eurasia. We also should note that Neanderthals also used Mousterian tools.
The Boule and Vallois research makes it clear that the Bushman expanded across Africa on into Europe via Spain as the Grimaldi people. This makes it clear that the Bushman/Khoisan people were not isolated in South Africa. The Khoisan people carry the haplogroup N. The Hadza are Bushman they carry haplogroup N.

The Aurignacian civilization was founded by the Cro-Magnon people who originated in Africa. They took this culture to Western Europe across the Straits of Gibraltar. The Cro-Magnon people were probably Bushman/Khoi.
There have been numerous "Negroid skeletons" found in Europe. Marcellin Boule and Henri Vallois, in Fossil Man, provide an entire chapter on the Africans/Negroes of Europe Anta Diop also discussed the Negroes of Europe inCivilization or Barbarism, pp.25-68. Also W.E. B. DuBois, discussed these Negroes in the The World and Africa, pp.86-89. DuBois noted that "There was once a an "uninterrupted belt' of Negro culture from Central Europe to South Africa" (p.88).
Boule and Vallois, note that "To sum up, in the most ancient skeletons from the Grotte des Enfants we have a human type which is readily comparable to modern types and especially to the Negritic or Negroid type" (p.289). They continue, "Two Neolithic individuals from Chamblandes in Switzerland are Negroid not only as regards their skulls but also in the proportions of their limbs. Several Ligurian and Lombard tombs of the Metal Ages have also yielded evidences of a Negroid element.
Since the publication of Verneau's memoir, discoveries of other Negroid skeletons in Neolithic levels in Illyria and the Balkans have been announced. The prehistoric statues, dating from the Copper Age, from Sultan Selo in Bulgaria are also thought to protray Negroids.
In 1928 Rene Bailly found in one of the caverns of Moniat, near Dinant in Belgium, a human skeleton of whose age it is difficult to be certain, but seems definitely prehistoric. It is remarkable for its Negroid characters, which give it a reseblance to the skeletons from both Grimaldi and Asselar (p.291).
Boule and Vallois, note that "We know now that the ethnography of South African tribes presents many striking similarities with the ethnography of our populations of the Reindeer Age. Not to speak of their stone implements which, as we shall see later , exhibit great similarities, Peringuey has told us that in certain burials on the South African coast 'associated with the Aurignacian or Solutrean type industry...."(p.318-319). They add, that in relation to Bushman art " This almost uninterrupted series leads us to regard the African continent as a centre of important migrations which at certain times may have played a great part in the stocking of Southern Europe. Finally, we must not forget that the Grimaldi Negroid skeletons sho many points of resemblance with the Bushman skeletons". They bear no less a resemblance to that of the fossil Man discovered at Asslar in mid-Sahara, whose characters led us to class him with the Hottentot-Bushman group.
The Boule and Vallois research makes it clear that the Bushman expanded across Africa on into Europe via Spain as the Grimaldi people. This makes it clear that the Bushman/Khoisan people were not isolated in South Africa. The Khoisan people carry the haplogroup N. The Hadza are Bushman they carry haplogroup N.
Cro-Magnon people carried haplogroup N:
quote:This suggest that haplogroup N was taken to Western Eurasia by the San people=Cro-Magnon.
Specific mtDNA sites outside HVRI were also analyzed (by amplification, cloning, and sequencing of the surrounding region) to classify more precisely the ancient sequences within the phylogenetic network of present-time mtDNAs (35, 36). Paglicci-25 has the following motifs: +7,025 AluI, 00073A, 11719G, and 12308A. Therefore, this sequence belongs to either haplogroups HV or pre-HV, two haplogroups rare in general but with a comparatively high frequencies among today's Near-Easterners (35). Paglicci-12 shows the motifs 00073G, 10873C, 10238T, and AACC between nucleotide positions 10397 and 10400, which allows the classification of this sequence into the macrohaplogroupN,containing haplogroups W, X, I, N1a, N1b, N1c, and N*. Following the definition given in ref. 36, the presence of a single mutation in 16,223 within HRVI suggests a classification of Paglicci-12 into the haplogroup N*, which is observed today in several samples from the Near East and, at lower frequencies, in the Caucasus (35). It is difficult to say whether the apparent evolutionary relationship between Paglicci-25 and Paglicci-12 and those populations is more than a coincidence. Indeed, the haplogroups to which the Cro-Magnon type sequences appear to belong are rare among modern samples, and therefore their frequencies are poorly estimated. However, genetic affinities between the first anatomically modern Europeans and current populations of the Near East make sense in the light of the likely routes of Upper Paleolithic human expansions in Europe, as documented in the archaeological record (37).
http://www.pnas.org/cgi/content/full/100/11/6593

In summary, the Black Berbers took African mtDNA into Western Europe over 40kya . in addition, Berbers and Khoisan continue to carry y-Chromosome haplogroups A1 and R1-M343. The Tuareg probably helped spread hg H in Europe after they invaded Europe along with other sahelians/Moors during the Islamic period.
- References:
Bekada A, Arauna LR, Deba T, Calafell F, Benhamamouch S, Comas D (2015) Genetic Heterogeneity in Algerian Human Populations. PLoS ONE 10(9): e0138453. doi:10.1371/journal.pone.0138453 http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0138453
Cruciani, F., Trombetta, B., Massaia, A., Destro-Bisol, G., Sellitto, D., & Scozzari, R. (2011). A Revised Root for the Human Y Chromosomal Phylogenetic Tree: The Origin of Patrilineal Diversity in Africa. American Journal of Human Genetics, 88(6), 814–818. http://doi.org/10.1016/j.ajhg.2011.05.002
Domínguez E.F. (2005). Polimorfismos de DNA mitocondrial en poblaciones antiguas de la cuenca
mediterránea. Universitat de Barcelona. Departament de Biologia Animal, 2005 (PhD thesis).
Frigi et al. (2010). Ancient Local Evolution of African mtDNA Haplogroups in Tunisian Berber Populations, Human Biology (August 2010 (82:4).
Rito T, Richards MB, Fernandes V, Alshamali F, Cerny V, et al. (2013) The First Modern Human Dispersals across Africa. PLoS ONE 8(11): e80031. doi:10.1371/journal.pone.0080031
Schlebusch, C.M. (2010). Genetic variation in Khoisan-speaking populations from southern Africa. University of the Witwatersrand, Johannesburg (2010 PhD thesis).https://www.academia.edu/3426993/Genetic_variation_in_Khoisan-speaking_populations_from_southern_Africa
Winters,C. (2010). Origin and spread of the Haplogroup N. Bioresearch Bull (2010) 3:116-122.
Winters C.(2011). The Gibraltar Out of Africa Exit for Anatomically Modern Humans. WebmedCentral BIOLOGY 2011;2(10):WMC002319 doi: 10.9754/journal.wmc.2011.002319
Winters, C. (2014). The Hadza Are Related to the South African Khoisan. http://www.exposingblacktruth.org/the-hadza-are-related-to-the-south-african-khoisan/
Winters,C.(2012). There has been a Continuous Indigenous Sub-Saharan Presence in North Africa for 30ky. Comment: . http://olmec98.net/ContinuousEurope.pdf